Piece-wise exponential (Additive Mixed) Modeling Tools
ISCB41, 2020
Andreas Bender (@adibender),
Fabian Scheipl, David Rügamer, Philipp Kopper, Bernd Bischl, Helmut Küchenhoff
Department of Statistics, LMU Munich
The framework is general in the sense that
it supports different Survival Tasks
- right-censoring, left-truncation
- time-varying effects, time-varying features
- cumulative effects (weighted cumulative exposure, distributed lag models)
- competing risks, multi-state models
does not require specialized Software, can be applied
- across programming languages and
- using any algorithm that supports optimization of the Poisson Likelihood
\usepackageamsmath,amssymb,bm
\usepackageamsmath,amssymb,bm
Survival Analysis as Poisson Regression
Consider setting with right-censored data:
- we observe (ti,δi),i=1,…,n, where
- ti=min(Ti,Ci); Ti∼F⊥Ci∼G;Ti,Ci>0
- δi=I(Ti≤Ci)∈{0,1}
To approximate λ(t;xi)=exp(g(xi(t),t))PH=λ0(t)exp(x′iβ)
Consider setting with right-censored data:
- we observe (ti,δi),i=1,…,n, where
- ti=min(Ti,Ci); Ti∼F⊥Ci∼G;Ti,Ci>0
- δi=I(Ti≤Ci)∈{0,1}
To approximate λ(t;xi)=exp(g(xi(t),t))PH=λ0(t)exp(x′iβ)
- split the follow-up in J intervals (κj−1,κj],j=1,…,J
Consider setting with right-censored data:
- we observe (ti,δi),i=1,…,n, where
- ti=min(Ti,Ci); Ti∼F⊥Ci∼G;Ti,Ci>0
- δi=I(Ti≤Ci)∈{0,1}
To approximate λ(t;xi)=exp(g(xi(t),t))PH=λ0(t)exp(x′iβ)
split the follow-up in J intervals (κj−1,κj],j=1,…,J
assume piece-wise constant hazards: λ(t|xi(t))≡exp(g(xij,tj)):=λij, ∀t∈(κj−1,κj],
Consider setting with right-censored data:
- we observe (ti,δi),i=1,…,n, where
- ti=min(Ti,Ci); Ti∼F⊥Ci∼G;Ti,Ci>0
- δi=I(Ti≤Ci)∈{0,1}
To approximate λ(t;xi)=exp(g(xi(t),t))PH=λ0(t)exp(x′iβ)
split the follow-up in J intervals (κj−1,κj],j=1,…,J
assume piece-wise constant hazards: λ(t|xi(t))≡exp(g(xij,tj)):=λij, ∀t∈(κj−1,κj],
- Estimation using
- Piece-wise Exponential Model (PEM; e.g.: Laird, et al. (1981); Friedman (1982); Carstensen, et al. (2011))
- Piece-wise exponential Additive Mixed Models (PAMM, e.g.: Cai, et al. (2002); Kauermann (2005); Argyropoulos, et al. (2015); Bender, et al. (2018))
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
- define: δij={1ti∈(κj−1,κj]∧δi=10else
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
- define: δij={1ti∈(κj−1,κj]∧δi=10else, tij={ti−κj−1δij=1κj−κj−1else
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
- define: δij={1ti∈(κj−1,κj]∧δi=10else, tij={ti−κj−1δij=1κj−κj−1else, tj:=κj
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
- define: δij={1ti∈(κj−1,κj]∧δi=10else, tij={ti−κj−1δij=1κj−κj−1else, tj:=κj
General log-likelihood contribution:
ℓi=log(λ(ti;xi)δiS(ti;xi))=Ji∑j=1(δijlogλij−λijtij)
Working Assumption δijiid∼Po(μij=λijtij):
ℓi=log(Ji∏j=1f(δij))=Ji∑j=1δijlog(λij)+δijlog(tij)−λijtij
Competing risks setting with event types k∈{1,2}
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
→ estimate λ(t|x,k)=exp(f(x(t),t,k)), k∈{1,2}
Competing risks setting with event types k∈{1,2}
→ transform to PED using κ0=0,κ1=1,κ2=1.5,κ3=3
→ estimate λ(t|x,k)=exp(f(x(tj),tj,k)), ∀t∈(κj−1,κj], k∈{1,2}
PEM/GLM: λ(t)=λ0j=exp(β0j),∀t∈(κj−1,κj],j=1,…,J
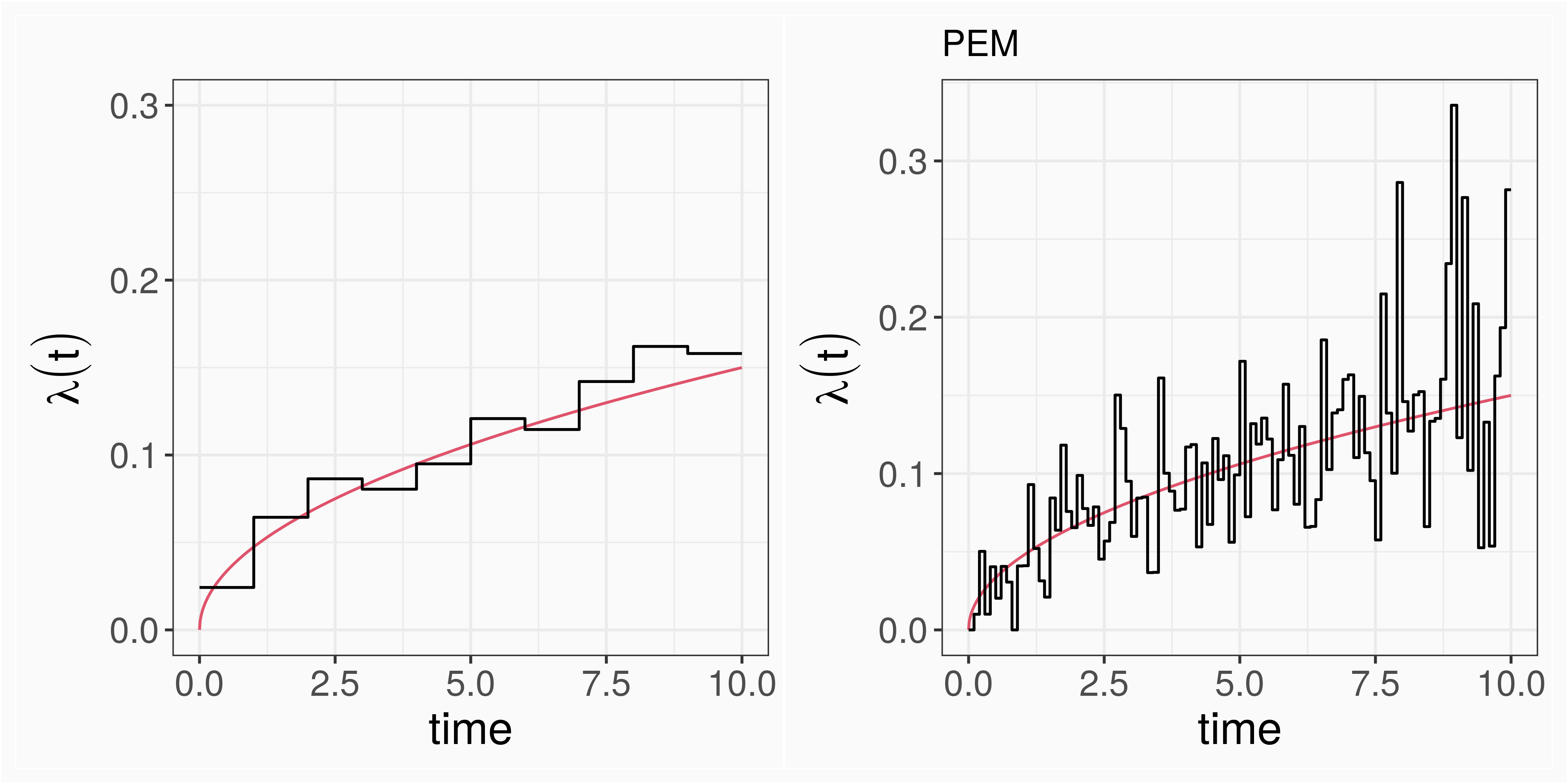
trade of w.r.t. to number of split points (less flexible/more robust vs. more flexible/less robust)
computationally inefficient (one parameter for each interval), especially when considering time-varying effects
results sensitive to number and placement of interval cut points
PAMM/GAMM: λ(t)=λ0j=exp(f0(tj)),∀t∈(κj−1,κj],j=1,…,J; f0(tj)=∑Qq=1β0qB0q(tj)
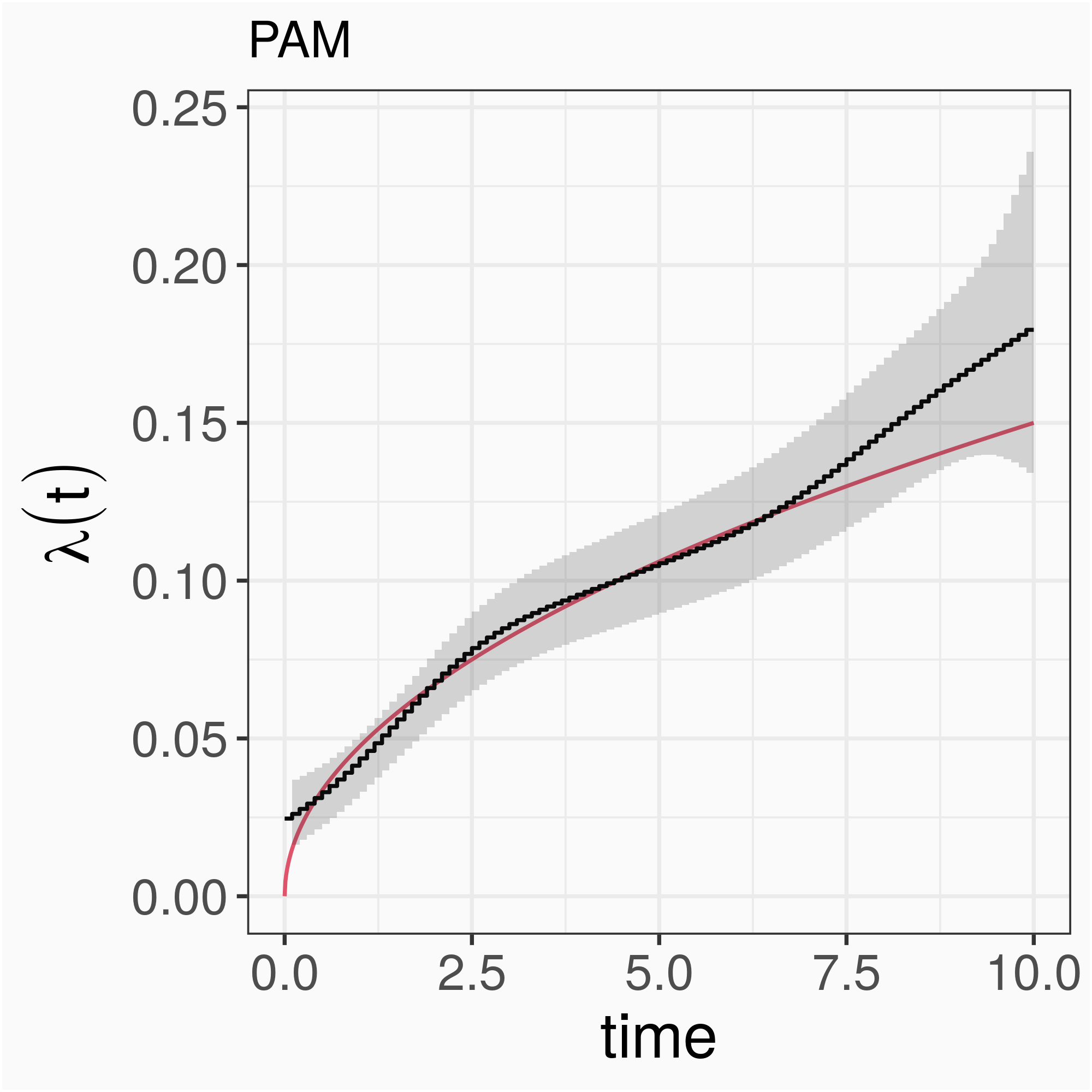
large differences between neighboring coefficients/baseline hazards of neighboring intervals are penalized
insensitive to number and placement of split points
number of parameters to estimate determined by basis dimension Q, not number of intervals J
Time-varying effects
In the PEM/PAMM framework, time-varying effects are simply interactions of
time tj and other covariates.
log(λ(t|x))=f01(tj)I(complications=yes)+f02(tj)I(complications=no)
pam_tumor <- mgcv::gam(formula=ped_status~s(tend, by=complications), data=ped_tumor, family=poisson(), offset=offset)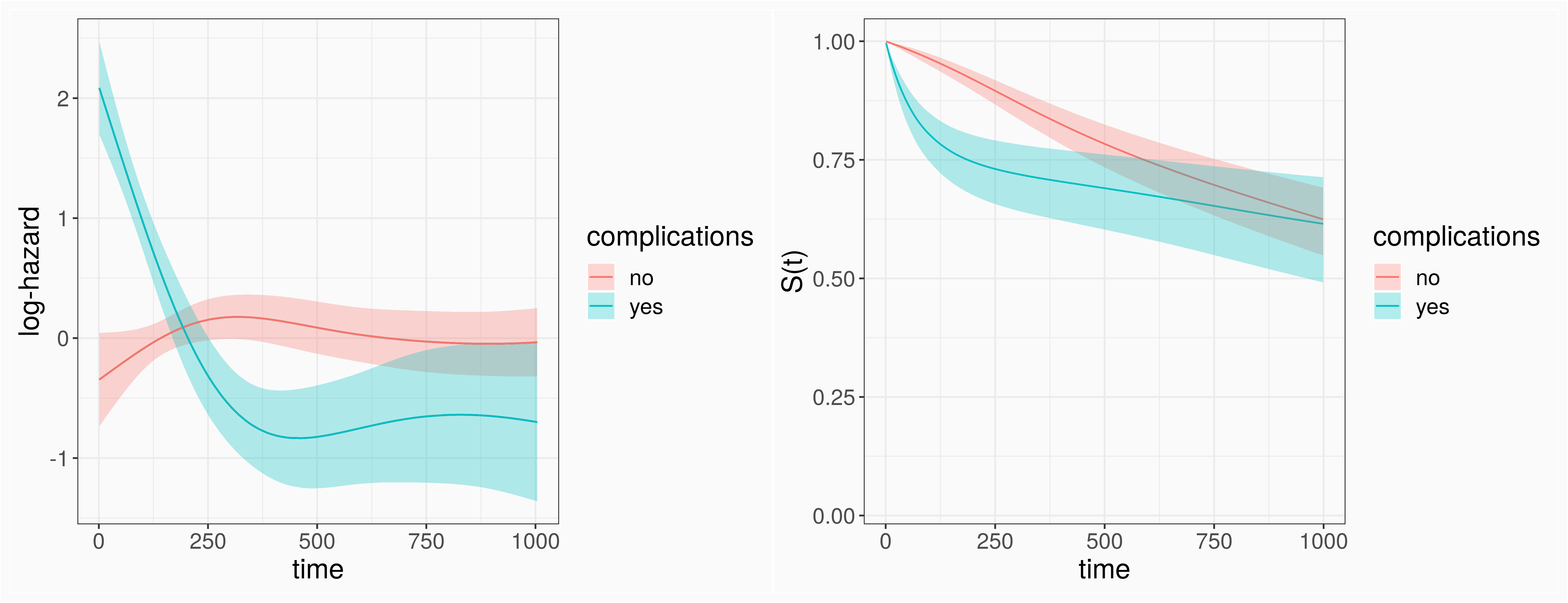
# "Regular" GAMmgcv::gam(formula=ped_status~s(tend, by=complications), data=ped_tumor, family=poisson(), offset=offset)# GAM with monotinicity constraintsscam::scam(formula=ped_status~s(tend, by=complications, bs = "mpd"), data=ped_tumor, family=poisson(), offset=offset)# Bayesian GAMbrms::brm(formula=ped_status~s(tend, by=complications) + offset(offset), data=ped_tumor, family=poisson())Competing Risks
log(λ(t|x))=f01(tj)I(k=1)+f01(tj)I(k=2) Cause specific hazards are time-varying effects of time tj and covariate "event type" k
pam_cr <- mgcv::gam(formula = ped_status ~ s(tend, by = cause), data = ped_stacked, family = poisson(), offset = offset)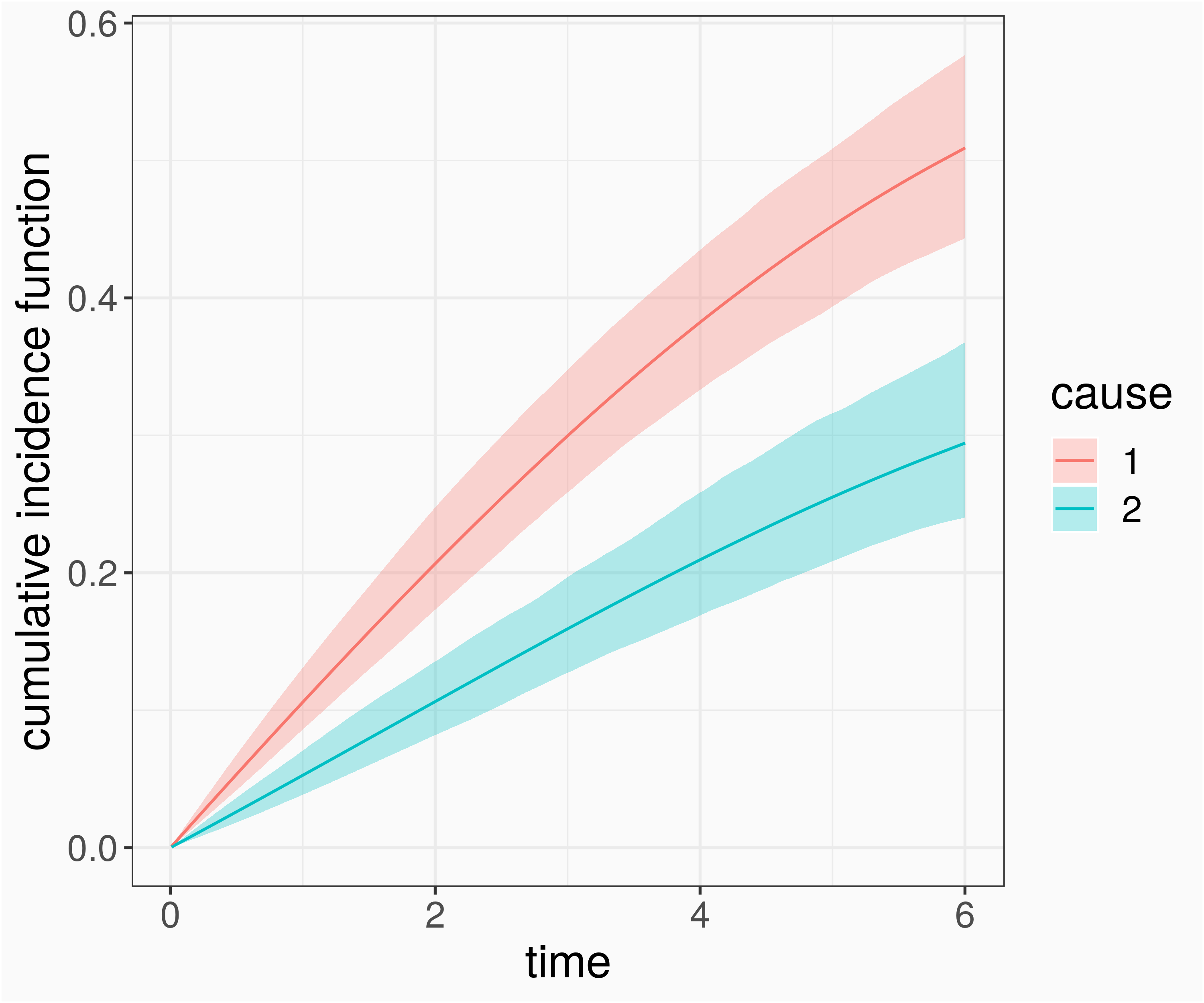
Tree based methods
Time-varying effects Shared vs. cause-specific effects (in CR)
(source: Bender, et al. (2020))
The pammtools package
PEMs/PAMMs powerfull framework for survival analysis, but cumbersome to work with
pammtools facilitates
data transformation (
as_ped):- right-censoring
- cumulative effects
- competing risks
post-processing:
- prediction (
add_hazard,add_surv_prob,add_cif), - model evaluation (integrated brier score via
pec)
- prediction (
convenience functions for visualisation, ...
Outlook
support for multi-state models
facilitate extensions: S3 functions for calculation of hazard for other packages (e.g.
mbooost,brms)Prototype for PEMs using
xgboostavailable: https://github.com/adibender/pem.xgbHowever, ML algorithms need a different infrastructure (resampling, tuning, benchmarking)
→ Development will probably continue inmlr3andmlr3proba(Lang, et al. (2019); Sonabend, et al. (2020))
References
Argyropoulos, C. et al. (2015). "Analysis of Time to Event Outcomes in Randomized Controlled Trials by Generalized Additive Models". In: PLoS ONE 10.4, p. e0123784. DOI: 10.1371/journal.pone.0123784. URL: http://dx.doi.org/10.1371/journal.pone.0123784.
Bender, A. et al. (2018). "A generalized additive model approach to time-to-event analysis". En. In: Statistical Modelling 18.3-4, pp. 299-321. ISSN: 1471-082X. DOI: 10.1177/1471082X17748083.
Bender, A. et al. (2020). "A General Machine Learning Framework for Survival Analysis". In: arXiv:2006.15442 [cs, stat]. arXiv: 2006.15442.
Cai, T. et al. (2002). "Mixed Model-Based Hazard Estimation". In: Journal of Computational and Graphical Statistics 11.4, pp. 784-798. ISSN: 1061-8600. DOI: 10.1198/106186002862. URL: http://dx.doi.org/10.1198/106186002862.
Carstensen, B. et al. (2011). "Using Lexis Objects for Multi-State Models in R". En. In: Journal of Statistical Software 38.1. Number: 1, pp. 1-18. ISSN: 1548-7660. DOI: 10.18637/jss.v038.i06. URL: https://www.jstatsoft.org/index.php/jss/article/view/v038i06.
Friedman, M. (1982). "Piecewise Exponential Models for Survival Data with Covariates". In: The Annals of Statistics 10.1, pp. 101-113. ISSN: 00905364. URL: http://www.jstor.org/stable/2240502.
References
Kauermann, G. (2005). "Penalized spline smoothing in multivariable survival models with varying coefficients". In: Computational Statistics & Data Analysis 49.1, pp. 169-186. ISSN: 0167-9473. DOI: 10.1016/j.csda.2004.05.006. URL: http://www.sciencedirect.com/science/article/pii/S0167947304001240.
Laird, N. et al. (1981). "Covariance Analysis of Censored Survival Data Using Log-Linear Analysis Techniques". In: Journal of the American Statistical Association 76.374, pp. 231-240. DOI: 10.2307/2287816. URL: http://www.jstor.org/stable/2287816.
Lang, M. et al. (2019). "mlr3: A modern object-oriented machine learning framework in R". In: Journal of Open Source Software. DOI: 10.21105/joss.01903. URL: https://joss.theoj.org/papers/10.21105/joss.01903.
Sonabend, R. et al. (2020). "mlr3proba: Machine Learning Survival Analysis in R". In: arXiv:2008.08080 [cs, stat]. arXiv: 2008.08080. URL: http://arxiv.org/abs/2008.08080 (visited on Aug. 20, 2020).