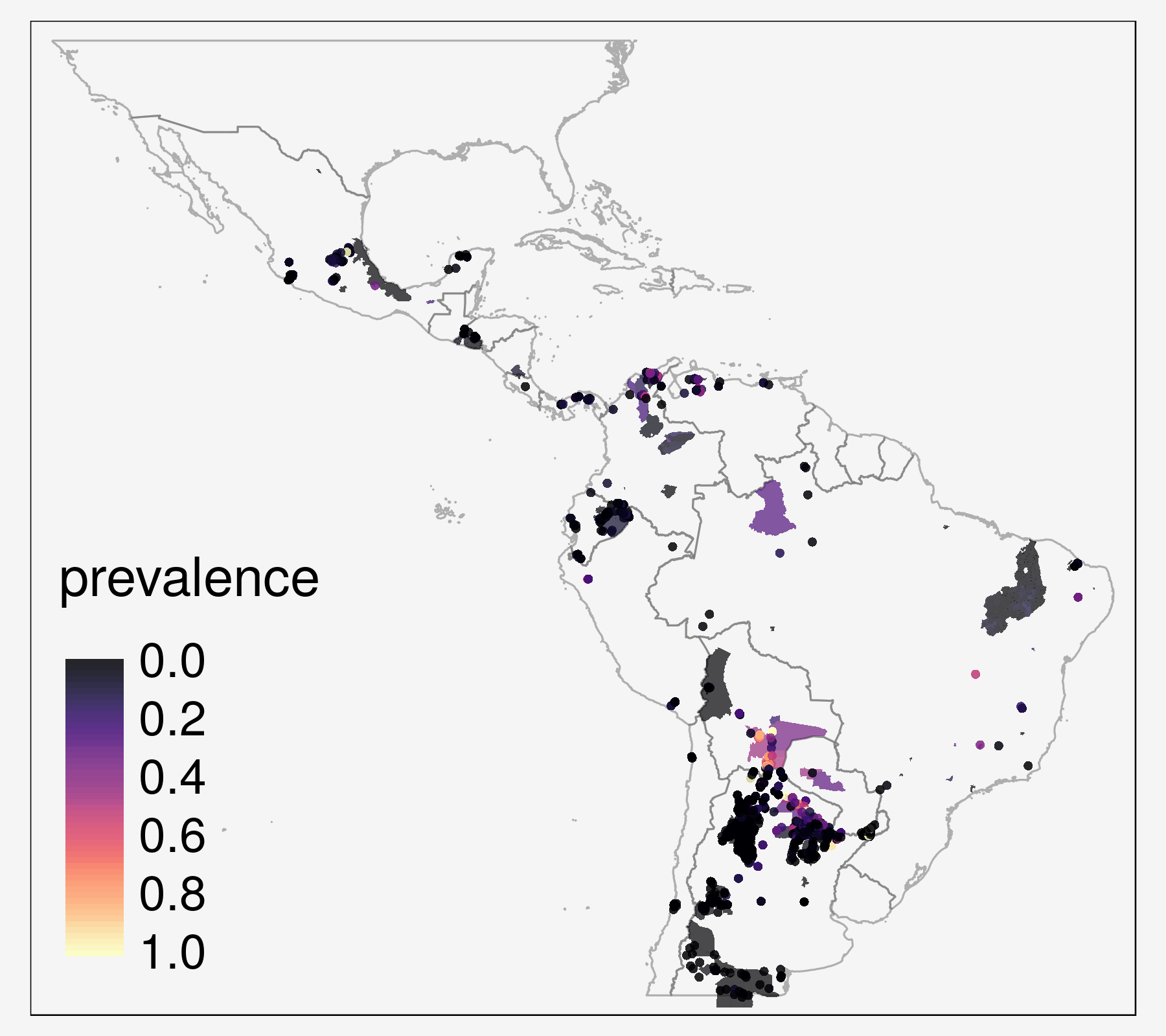
2. Infection prevalence in vectors
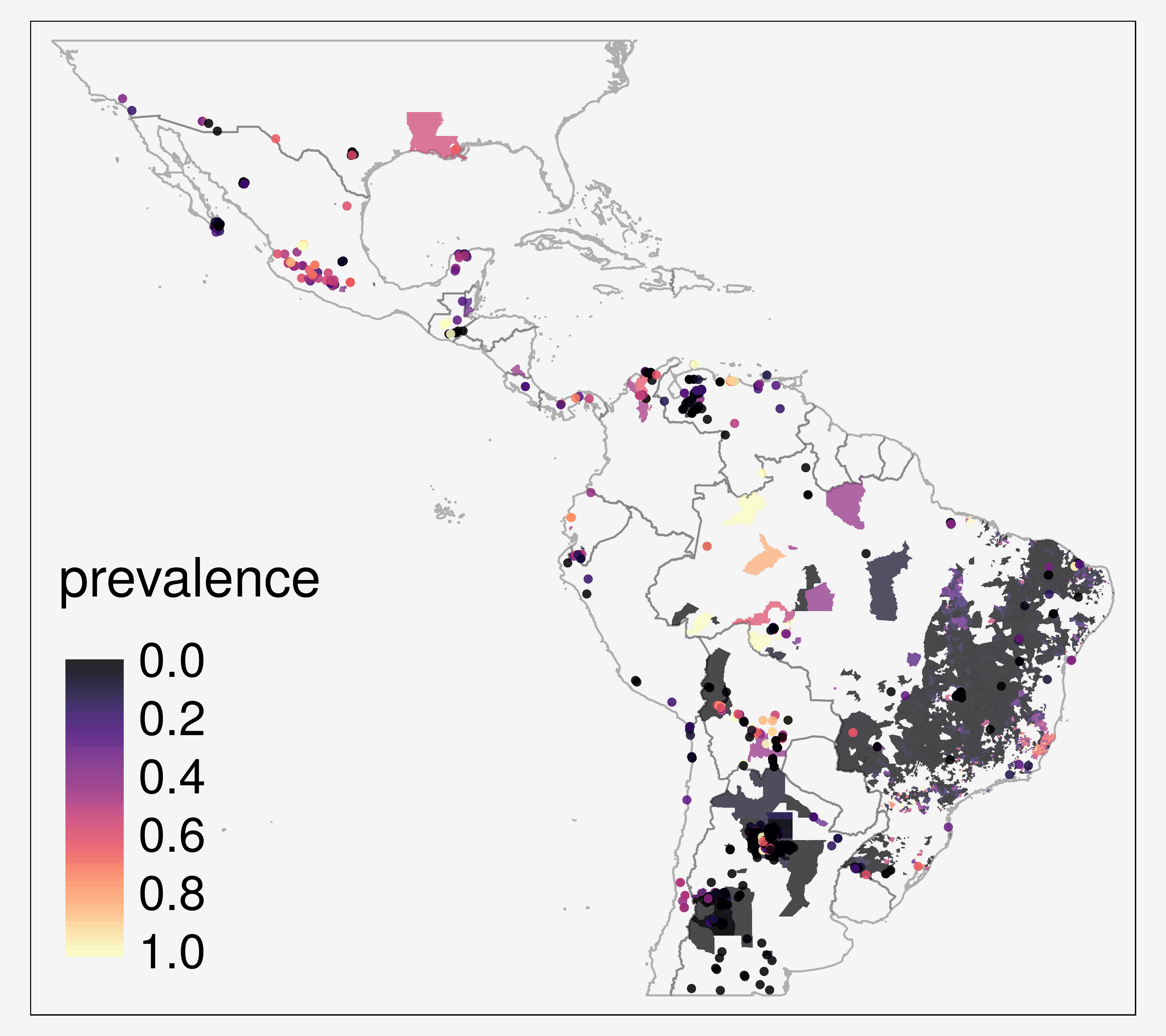
- point- and
polygon-level data - spatial sparseness
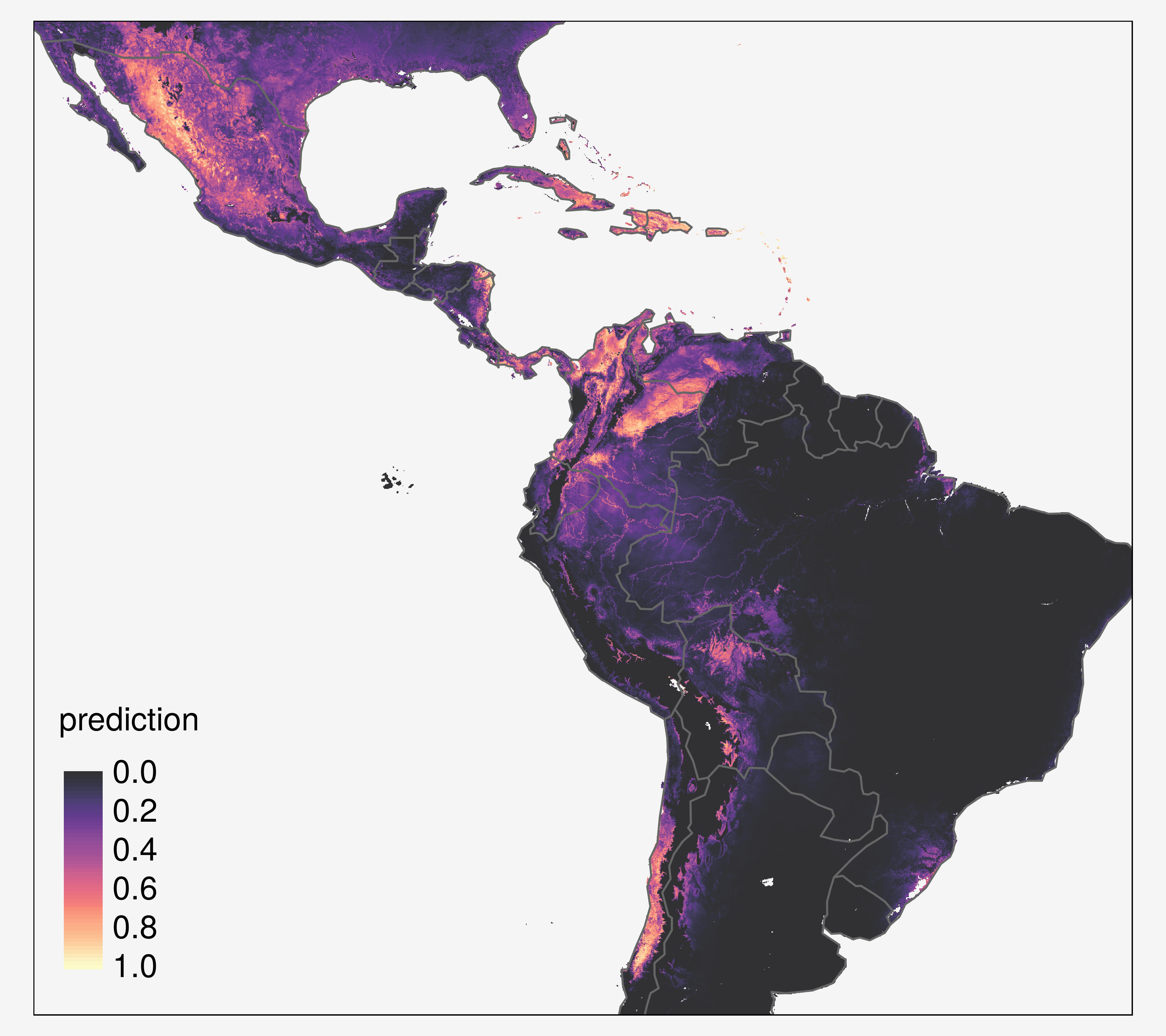
American trypanosomiasis, or Chagas disease is a leading cause of heart disease in Latin America
It is one of the 10 neglected diseases addressed by the London Declaration which calls for control and elimination of these devastating diseases by 2020, based on the World Health Organization (WHO) Road map for overcoming the global impact of neglected tropical diseases.
Caused by infection with the Trypanosoma cruzi parasite
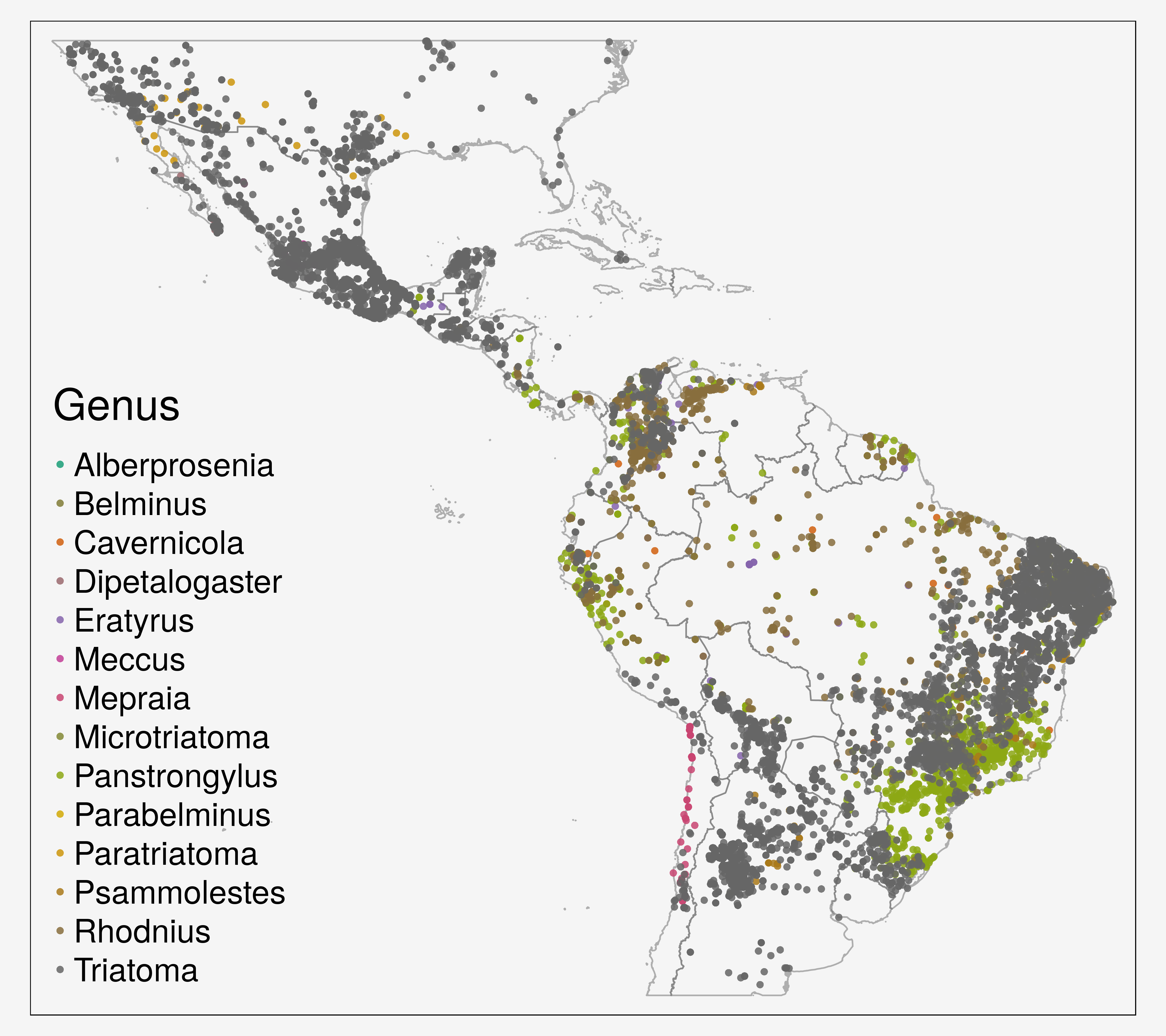
Transmitted to humans by triatomine vectors (100 species from 18 different genera)
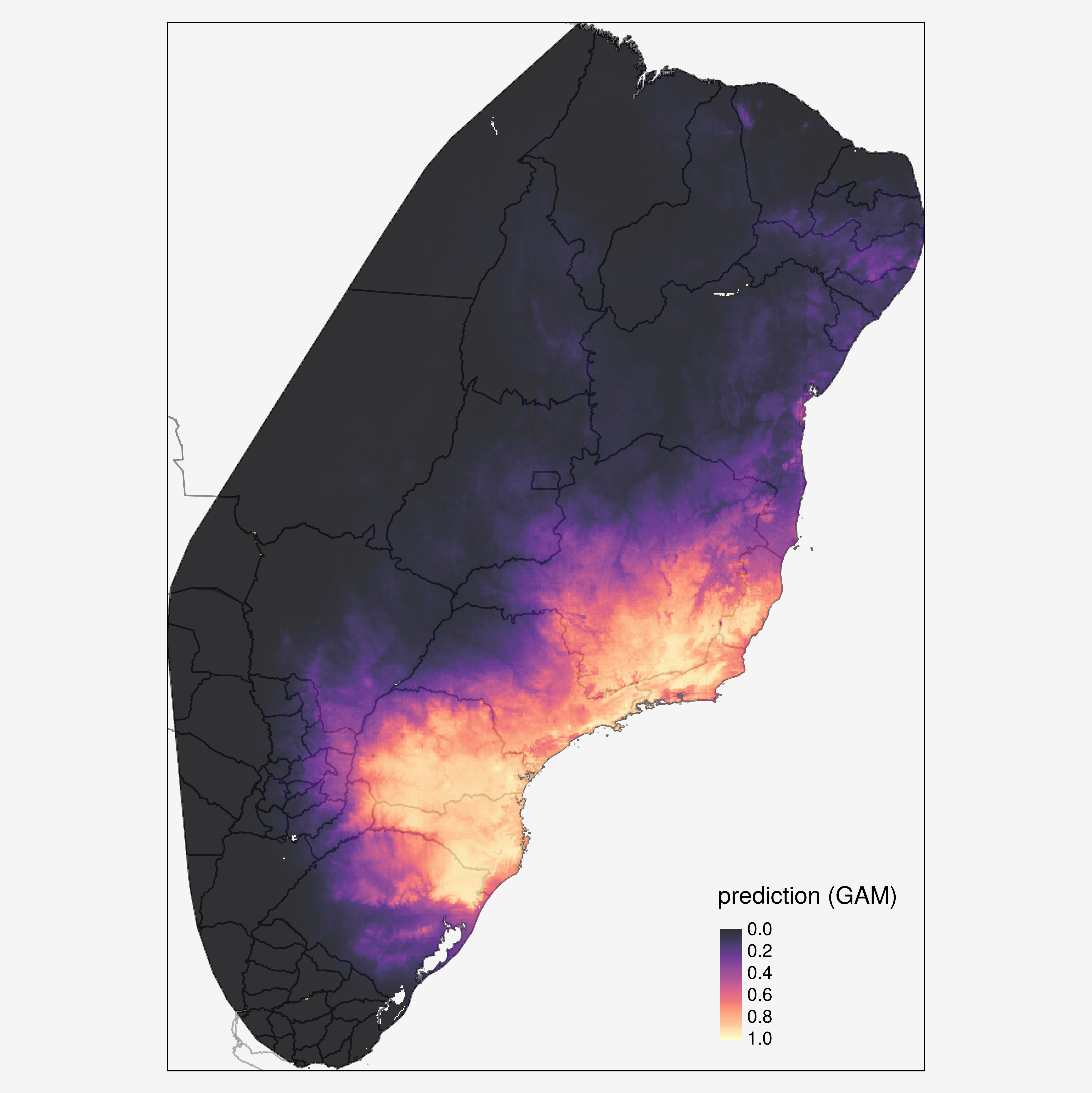
The primary aim of this project is to map the prevalence of T. cruzi infections in vector species, in order to investigate spatial heterogeneity in vectorial transmission, to target vector control, and to quantify the population at risk.







η(s)=P∑p=1fp(xp(s))+GP(s); p(s)=(1−exp(η(s)))−1
η(s)=P∑p=1fp(xp(s))+GP(s); p(s)=(1−exp(η(s)))−1
η(s)=P∑p=1fp(xp(s))+GP(s); p(s)=(1−exp(η(s)))−1







How much does outdoor biting contribute to transmission?
How much does contamination of food/drink by vectors contribute to transmission?
How long after the elimination of vectors from homes (through insecticide spraying) will re-infestation occur?
Does insecticide resistance have the potential to derail the elimination of vectors from homes?
Does the effect of improved housing change if all houses are improved, compared to the situation where there is a mosaic of poor-quality and higher-quality housing?
Bhatt, S, E. Cameron, S. R. Flaxman, et al. (2017). "Improved prediction accuracy for disease risk mapping using Gaussian process stacked generalization". En. In: Journal of The Royal Society Interface 14.134, p. 20170520. DOI: 10.1098/rsif.2017.0520.
Browne, A. J, C. A. Guerra, R. V. Alves, et al. (2017). "The contemporary distribution of Trypanosoma cruzi infection in humans, alternative hosts and vectors". En. In: Scientific Data 4, p. 170050. DOI: 10.1038/sdata.2017.50.
Ceccarelli, S, A. Balsalobre, P. Medone, et al. (2018). "DataTri, a database of American triatomine species occurrence". En. In: Scientific Data 5, p. 180071. DOI: 10.1038/sdata.2018.71.
Miller, D. A. W, K. Pacifici, J. S. Sanderlin, et al. (2019). "The recent past and promising future for data integration methods to estimate species’ distributions". En. In: Methods in Ecology and Evolution 10.1, pp. 22-37. DOI: 10.1111/2041-210X.13110.
Phillips, S. J, M. Dudík, J. Elith, et al. (2009). "Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data". En. In: Ecological Applications 19.1, pp. 181-197. DOI: 10.1890/07-2153.1.
Roberts, D. R, V. Bahn, S. Ciuti, et al. (2017). "Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure". En. In: Ecography 40.8, pp. 913-929. DOI: 10.1111/ecog.02881.
Valavi, R, J. Elith, J. J. Lahoz-Monfort, et al. (2018). "blockCV: An r package for generating spatially or environmentally separated folds for k-fold cross-validation of species distribution models". In: Methods in Ecology and Evolution 0.0. DOI: 10.1111/2041-210X.13107.
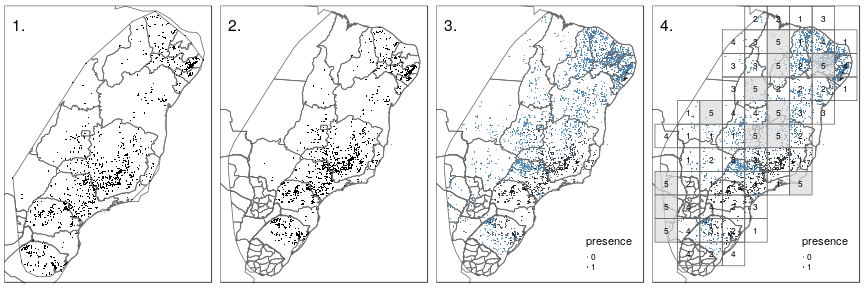
Convex hull around species presence locations
Extended convex hull
Define absences as reported presence locations of all other species
Set up spatial block-wise cross-validation (each fold 1-5 ∼ same proportion of presences and absences)
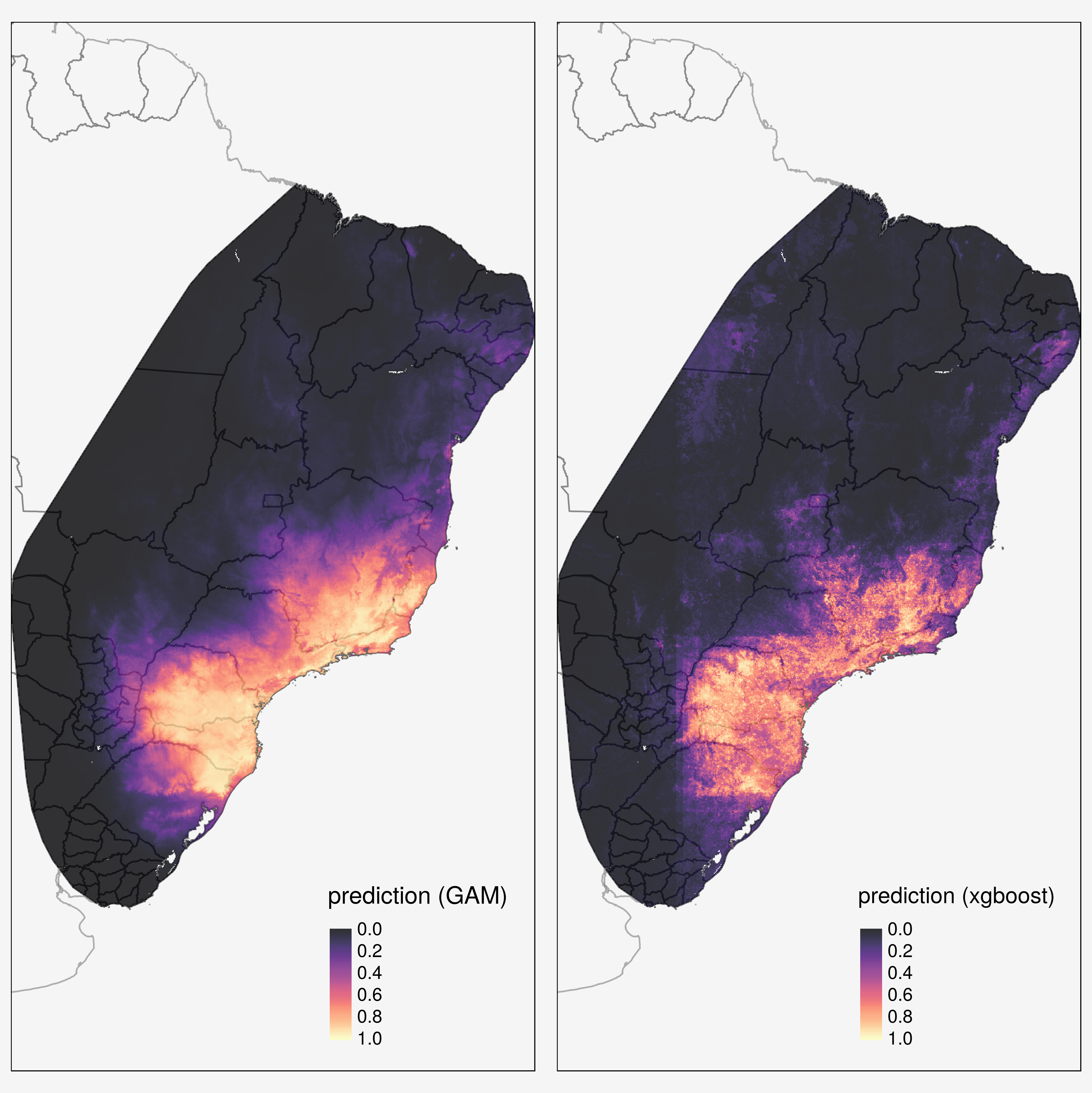
American trypanosomiasis, or Chagas disease is a leading cause of heart disease in Latin America
It is one of the 10 neglected diseases addressed by the London Declaration which calls for control and elimination of these devastating diseases by 2020, based on the World Health Organization (WHO) Road map for overcoming the global impact of neglected tropical diseases.
Caused by infection with the Trypanosoma cruzi parasite
Transmitted to humans by triatomine vectors (100 species from 18 different genera)
The primary aim of this project is to map the prevalence of T. cruzi infections in vector species, in order to investigate spatial heterogeneity in vectorial transmission, to target vector control, and to quantify the population at risk.
Keyboard shortcuts
| ↑, ←, Pg Up, k | Go to previous slide |
| ↓, →, Pg Dn, Space, j | Go to next slide |
| Home | Go to first slide |
| End | Go to last slide |
| Number + Return | Go to specific slide |
| b / m / f | Toggle blackout / mirrored / fullscreen mode |
| c | Clone slideshow |
| p | Toggle presenter mode |
| t | Restart the presentation timer |
| ?, h | Toggle this help |
| Esc | Back to slideshow |